A Tour of Xarray Customizations#
Xarray is easily extensible. This means it is easy to add on to to build custom packages that tackle particular computational problems or supply domain specific functionality.
These packages can plug in to xarray in various different ways. They may build directly on top of xarray, or they may take advantage of some of xarray’s dedicated interfacing features:
Accessors (extensions)
Backend (filetype) entrypoint
Metadata attributes
Duck-array wrapping interface
Flexible indexes (coming soon!)
Here we introduce several popular or interesting extensions that are installable as their own packages (via conda and pip). These packages integrate with xarray using one or more of the features mentioned above.
hvplot, a powerful interactive plotting library
rioxarray, for working with geospatial raster data using rasterio
cf-xarray, for interpreting CF-compliant data
pint-xarray, for unit-aware computations using pint.
Specific examples for implementing your own xarray customizations using these interfacing features are available in the ADVANCED section of this book.
Quick note on accessors#
Before we look at the packages we need to briefly introduce a feature they commonly use: “xarray accessors”.
The accessor-style syntax is used heavily by the other libraries we are about to cover.
For users, accessors just allow us to have a familiar dot (method-like) syntax on xarray objects, for example da.hvplot(), da.pint.quantify(), or da.cf.describe().
hvplot via accessors#
The HoloViews library makes great use of accessors to allow seamless plotting of xarray data using a completely different plotting backend (by default, xarray uses matplotlib).
We first need to import the code that registers the hvplot accessor
import hvplot.xarray
And now we can call the .hvplot method to plot using holoviews in the same way that we would have used .plot to plot using matplotlib.
ds = xr.tutorial.load_dataset("air_temperature")
ds['air'].isel(time=1).hvplot(cmap="fire")
For some more examples of how powerful HoloViews is see here.
Rioxarray via the backend entrypoint#
Rioxarray is a Python library that enhances Xarray’s ability to work with geospatial data and coordinate reference systems. Geographic information systems use GeoTIFF and many other formats to organize and store gridded, or raster, datasets.
The Geospatial Data Abstraction Library (GDAL) provides foundational drivers and geospatial algorithms, and the rasterio library provides a Pythonic interface to GDAL. Rioxarray brings key features of rasterio to Xarray:
A backend engine to read any format recognized by GDAL
A
.rioaccessor for rasterio’s geospatial algorithms such as reprojection
Below a couple brief examples to illustrate these features:
import rioxarray # ensure you have rioxarray installed in your environment
You can explicitly use rioxarray’s ‘rasterio’ engine to load myriad geospatial raster formats. Below is a Cloud-Optimized Geotiff from an AWS public dataset of synthetic aperture radar data over Washington, State, USA. overview_level=4 is an argument specific to the rasterio engine that allows opening a pre-computed lower resolution “overview” of the data.
url = 'https://sentinel-s1-rtc-indigo.s3.us-west-2.amazonaws.com/tiles/RTC/1/IW/10/U/CU/2017/S1A_20170101_10UCU_ASC/Gamma0_VV.tif'
da = xr.open_dataarray(url, engine='rasterio', open_kwargs={"overview_level": 2})
da
<xarray.DataArray 'band_data' (band: 1, y: 687, x: 687)> Size: 2MB
[471969 values with dtype=float32]
Coordinates:
* band (band) int64 8B 1
* x (x) float64 5kB 3.001e+05 3.002e+05 ... 4.096e+05 4.097e+05
* y (y) float64 5kB 5.4e+06 5.4e+06 5.4e+06 ... 5.29e+06 5.29e+06
spatial_ref int64 8B ...
Attributes:
ABSOLUTE_ORBIT_NUMBER: 14632
DATE: 2017-01-01
MISSION_ID: S1A
NUMBER_SCENES: 1
ORBIT_DIRECTION: ascending
OVR_RESAMPLING_ALG: AVERAGE
SCENES: S1A_IW_GRDH_1SDV_20170101T021007_20170101T021036_...
SCENE_1_METADATA: {"title": "S1A_IW_GRDH_1SDV_20170101T021007_20170...
SCENE_1_PRODUCT_INFO: {"id": "S1A_IW_GRDH_1SDV_20170101T021007_20170101...
TILE_ID: 10UCU
VALID_PIXEL_PERCENT: 66.318
AREA_OR_POINT: AreaThe spatial_ref coordinate is added by rioxarray to store standardized geospatial Coordinate Reference System (CRS) information. We can access that information and additional methods via the .rio accessor:
da.rio.crs
CRS.from_wkt('PROJCS["WGS 84 / UTM zone 10N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",-123],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32610"]]')
EPSG refers to ‘European Petroleum Survey Group’, a database of the many CRS definitions for our Planet used over the years! EPSG=32610 is the “UTM 10N” CRS, with coordinate units in meters. Let’s say you want longitude,latitude coordinate points in degrees instead. You’d have to reproject this data:
da_lonlat = da.rio.reproject('epsg:4326')
da_lonlat
<xarray.DataArray 'band_data' (band: 1, y: 548, x: 820)> Size: 2MB
array([[[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]]],
shape=(1, 548, 820), dtype=float32)
Coordinates:
* x (x) float64 7kB -125.7 -125.7 -125.7 ... -124.2 -124.2 -124.2
* y (y) float64 4kB 48.75 48.74 48.74 48.74 ... 47.74 47.74 47.73
* band (band) int64 8B 1
spatial_ref int64 8B 0
Attributes:
ABSOLUTE_ORBIT_NUMBER: 14632
DATE: 2017-01-01
MISSION_ID: S1A
NUMBER_SCENES: 1
ORBIT_DIRECTION: ascending
OVR_RESAMPLING_ALG: AVERAGE
SCENES: S1A_IW_GRDH_1SDV_20170101T021007_20170101T021036_...
SCENE_1_METADATA: {"title": "S1A_IW_GRDH_1SDV_20170101T021007_20170...
SCENE_1_PRODUCT_INFO: {"id": "S1A_IW_GRDH_1SDV_20170101T021007_20170101...
TILE_ID: 10UCU
VALID_PIXEL_PERCENT: 66.318
AREA_OR_POINT: AreaNote that that the size of the data has changed as well as the coordinate values. This is typical of reprojection, as your data must be resampled and often interpolated to match the new CRS grid! A quick plot will compare the results of our reprojected data:
import panel as pn
img1 = da.sel(band=1).hvplot.image(
x='x', y='y', rasterize=True, cmap='gray', clim=(0, 0.5), title='UTM'
)
img2 = da_lonlat.sel(band=1).hvplot.image(
rasterize=True, cmap='gray', clim=(0, 0.5), title='LON/LAT'
)
pn.Column(img1, img2)
cf-xarray via metadata attributes#
Xarray objects can store arbitrary metadata in the form of a dict attached to each DataArray and Dataset object, accessible via the .attrs property.
xr.DataArray(name="Hitchhiker", data=0, attrs={"life": 42, "name": "Arthur Dent"})
<xarray.DataArray 'Hitchhiker' ()> Size: 8B
array(0)
Attributes:
life: 42
name: Arthur DentNormally xarray operations ignore this metadata, simply carting it around until you explicitly choose to use it. However sometimes we might want to write custom code which makes use of the metadata.
cf_xarray is a project that tries to
let you make use of other Climate and Forecast metadata convention attributes (or “CF attributes”) that xarray ignores. It attaches itself
to all xarray objects under the .cf namespace.
Where xarray allows you to specify dimension names for analysis, cf_xarray
lets you specify logical names like "latitude" or "longitude" instead as
long as the appropriate CF attributes are set.
For example, the "longitude" dimension in different files might be labelled as: (lon, LON, long, x…), but cf_xarray let’s you always refer to the logical name "longitude" in your code:
import cf_xarray
# describe cf attributes in dataset
ds.air.cf.describe()
Coordinates:
CF Axes: * X: ['lon']
* Y: ['lat']
* T: ['time']
Z: n/a
CF Coordinates: * longitude: ['lon']
* latitude: ['lat']
* time: ['time']
vertical: n/a
Cell Measures: area, volume: n/a
Standard Names: * latitude: ['lat']
* longitude: ['lon']
* time: ['time']
Bounds: n/a
Grid Mappings: n/a
/tmp/ipykernel_5797/2553772647.py:2: DeprecationWarning: 'obj.cf.describe()' will be removed in a future version. Use instead 'repr(obj.cf)' or 'obj.cf' in a Jupyter environment.
ds.air.cf.describe()
The following mean operation will work with any dataset that has appropriate
attributes set that allow detection of the “latitude” variable (e.g.
units: "degress_north" or standard_name: "latitude")
# demonstrate equivalent of .mean("lat")
ds.air.cf.mean("latitude")
<xarray.DataArray 'air' (time: 2920, lon: 53)> Size: 1MB
array([[279.398 , 279.6664, 279.6612, ..., 279.9508, 280.3152, 280.6624],
[279.0572, 279.538 , 279.7296, ..., 279.7756, 280.27 , 280.7976],
[279.0104, 279.2808, 279.5508, ..., 279.682 , 280.1976, 280.814 ],
...,
[279.63 , 279.934 , 280.534 , ..., 279.802 , 280.346 , 280.778 ],
[279.398 , 279.666 , 280.318 , ..., 279.766 , 280.342 , 280.834 ],
[279.27 , 279.354 , 279.882 , ..., 279.426 , 279.97 , 280.482 ]],
shape=(2920, 53))
Coordinates:
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00# demonstrate indexing
ds.air.cf.sel(longitude=242.5, method="nearest")
<xarray.DataArray 'air' (time: 2920, lat: 25)> Size: 584kB
array([[241. , 238. , 239.7 , ..., 292. , 293.9 , 296.79],
[240. , 238.39, 241.1 , ..., 292.6 , 294.1 , 296.7 ],
[240.7 , 238.89, 240.8 , ..., 292.29, 293.4 , 296.1 ],
...,
[241.79, 243.49, 246.49, ..., 294.69, 296.69, 298.49],
[239.89, 241.69, 242.29, ..., 295.09, 296.89, 298.59],
[239.59, 241.49, 240.79, ..., 295.19, 296.79, 298.89]],
shape=(2920, 25))
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
lon float32 4B 242.5
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]Pint via duck array wrapping#
Why use pint#
Pint defines physical units, allowing you to work with numpy-like arrays which track the units of your values through computations.
Pint defines a numpy-like array class called pint.Quantity, which is made up of a numpy array and a pint.Unit instance.
from pint import Unit
# you can create a pint.Quantity by multiplying a value by a pint.Unit
d = np.array(10) * Unit("metres")
d
These units are automatically propagated through operations
t = 1 * Unit("seconds")
v = d / t
v
Or if the operation involves inconsistent units, a pint.DimensionalityError is raised.
d + t
---------------------------------------------------------------------------
DimensionalityError Traceback (most recent call last)
Cell In[17], line 1
----> 1 d + t
File ~/work/xarray-tutorial/xarray-tutorial/.pixi/envs/default/lib/python3.12/site-packages/pint/facets/plain/quantity.py:849, in PlainQuantity.__add__(self, other)
846 if isinstance(other, datetime.datetime):
847 return self.to_timedelta() + other
--> 849 return self._add_sub(other, operator.add)
File ~/work/xarray-tutorial/xarray-tutorial/.pixi/envs/default/lib/python3.12/site-packages/pint/facets/plain/quantity.py:101, in check_implemented.<locals>.wrapped(self, *args, **kwargs)
99 elif isinstance(other, list) and other and isinstance(other[0], type(self)):
100 return NotImplemented
--> 101 return f(self, *args, **kwargs)
File ~/work/xarray-tutorial/xarray-tutorial/.pixi/envs/default/lib/python3.12/site-packages/pint/facets/plain/quantity.py:751, in PlainQuantity._add_sub(self, other, op)
748 return self.__class__(magnitude, units)
750 if not self.dimensionality == other.dimensionality:
--> 751 raise DimensionalityError(
752 self._units, other._units, self.dimensionality, other.dimensionality
753 )
755 # Next we define some variables to make if-clauses more readable.
756 self_non_mul_units = self._get_non_multiplicative_units()
DimensionalityError: Cannot convert from 'meter' ([length]) to 'second' ([time])
pint inside xarray objects#
We have already seen that xarray can wrap numpy arrays or dask arrays, but in fact xarray can wrap any type of array that behaves similarly to a numpy array.
Using this feature we can store a pint.Quantity array inside an xarray DataArray
da = xr.DataArray(d)
da
<xarray.DataArray ()> Size: 8B <Quantity(10, 'meter')>
We can see that the data type stored within the DataArray is a Quantity object, rather than just a np.ndarray object, and the units of the data are displayed in the repr.
The reason this works is that a pint.Quantity array is what we call a “duck array”, in that it behaves so similarly to a numpy.ndarray that xarray can treat them the same way. (See python duck typing)
pint-xarray#
The convenience package pint-xarray makes it easier to get the benefits of pint whilst working with xarray objects.
It provides utility accessor methods for promoting xarray data to pint quantities (which we call “quantifying”) in various ways and for manipulating the resulting objects.
# to be able to read unit attributes following the CF conventions
import cf_xarray.units
import pint_xarray
xr.set_options(display_expand_data=False)
<xarray.core.options.set_options at 0x7f398c9d8b60>
pint-xarray provides the .pint accessor, which firstly allows us to easily extract the units of our data
da.pint.units
The .pint.quantify() accessor gives us various ways to convert normal xarray data to be unit-aware.
Note
It is preferred to use .pint.quantify() to convert xarray data to use pint rather than explicitly creating a pint.Quantity array and placing it inside the xarray object, because pint-xarray will handle various subtleties involving dask etc.
We can explicitly specify the units we want
da = xr.DataArray([4.5, 6.7, 3.8], dims="time")
da.pint.quantify("V")
<xarray.DataArray (time: 3)> Size: 24B [V] 4.5 6.7 3.8 Dimensions without coordinates: time
Or if the data has a “units” entry in its .attrs metadata dictionary, we can automatically use that to convert each variable.
For example, the xarray tutorial dataset we opened earlier has units in its attributes
ds.air.attrs['units']
'degK'
which we can automatically read with .pint.quantify():
quantified_air = ds.pint.quantify()
quantified_air
<xarray.Dataset> Size: 31MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B [degrees_north] 75.0 72.5 70.0 ... 17.5 15.0
* lon (lon) float32 212B [degrees_east] 200.0 202.5 205.0 ... 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float64 31MB [K] 241.2 242.5 243.5 ... 296.2 295.7
Indexes:
lat PintIndex(PandasIndex)
lon PintIndex(PandasIndex)
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Here we have actually gone even further, and used cf-xarray to automatically interpret cf-compliant units in the .attrs to valid pint units.
This automatically happened just as a result of importing cf-xarray above.
When we plot quantified data with xarray the correct units will automatically appear on the plot
quantified_air["air"].isel(time=500).plot();
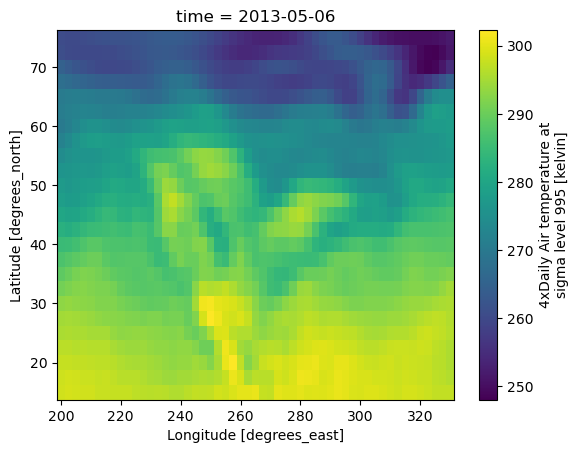
If we want to cast the pint arrays back to numpy arrays, we can use .pint.dequantify(), which will also write the current units back out to the .attrs["units"] field
quantified_air.pint.dequantify()
<xarray.Dataset> Size: 31MB
Dimensions: (time: 2920, lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float64 31MB 241.2 242.5 243.5 ... 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Exercise
Write a function which will raise a pint.DimensionalityError if supplied with Xarray DataArray with the wrong units.
Solution
from pint import DimensionalityError
def special_science_function(distance):
if distance.pint.units != "miles":
raise DimensionalityError(
"this function will only give the correct answer if the input is in units of miles"
)
Exercise
Try this on some of your data!
After you have imported pint-xarray (and maybe cf-xarray) as above, start with something like
ds = xr.open_dataset(my_data).pint.quantify()
Take a look at the pint-xarray documentation or the pint documentation if you get stuck.
The wider world…#
There are many other libraries in the wider xarray ecosystem. For a list of a few packages we particularly like for geoscience work see here, and for a more exhaustive list see here.

