Hierarchical storage formats#
In the fundamentals section we saw that xarray can read and write to a variety of storage formats. We have also seen that xarray’s data model can include a hierarchy of groups as part of an xarray.DataTree object.
The design of each format makes certain choices, and here we will compare the structure of common data formats to xarray’s full data model, concentrating on subtle differences.
Xarray is not a file format#
You sometimes hear people say things like “save it as an xarray”. This does not make sense, because xarray is not a file-format, it is an in-memory data structure. It’s analogous to the difference between a CSV file and a pandas.DataFrame.
Xarray data structures were inspired by scientific file formats (particularly netCDF), but are not intended to be identical to any of them. This helps xarray be a completely domain-agnostic tool.
Xarray supports reading from and writing to a range of file formats, but as common file formats have differences in their design, xarray data structures cannot be exactly equivalent to all of them.
Overall though this makes sense because the use case is different: in-memory data is for analysis, on-disk data is for persistent storage.
Groups everywhere#
Many storage formats for scientific data include some notion of “groups”.
The exact meaning of “group” differs between formats but are all ultimately motivated by a common recognition: that real scientific datasets often include related but otherwise heterogeneous data.
Zarr#
We’ll start with Zarr, because it has the most simple type of hierarchical structure.
Tree of groups – Tree of arbitrary groups.
Separate groups – No relationship enforced between groups, and no references from one group to another.
Separate arrays – No relationship enforced between arrays within a group.
Arbitrary JSON metadata – Each holds arbitrary data in the form of arrays + metadata.
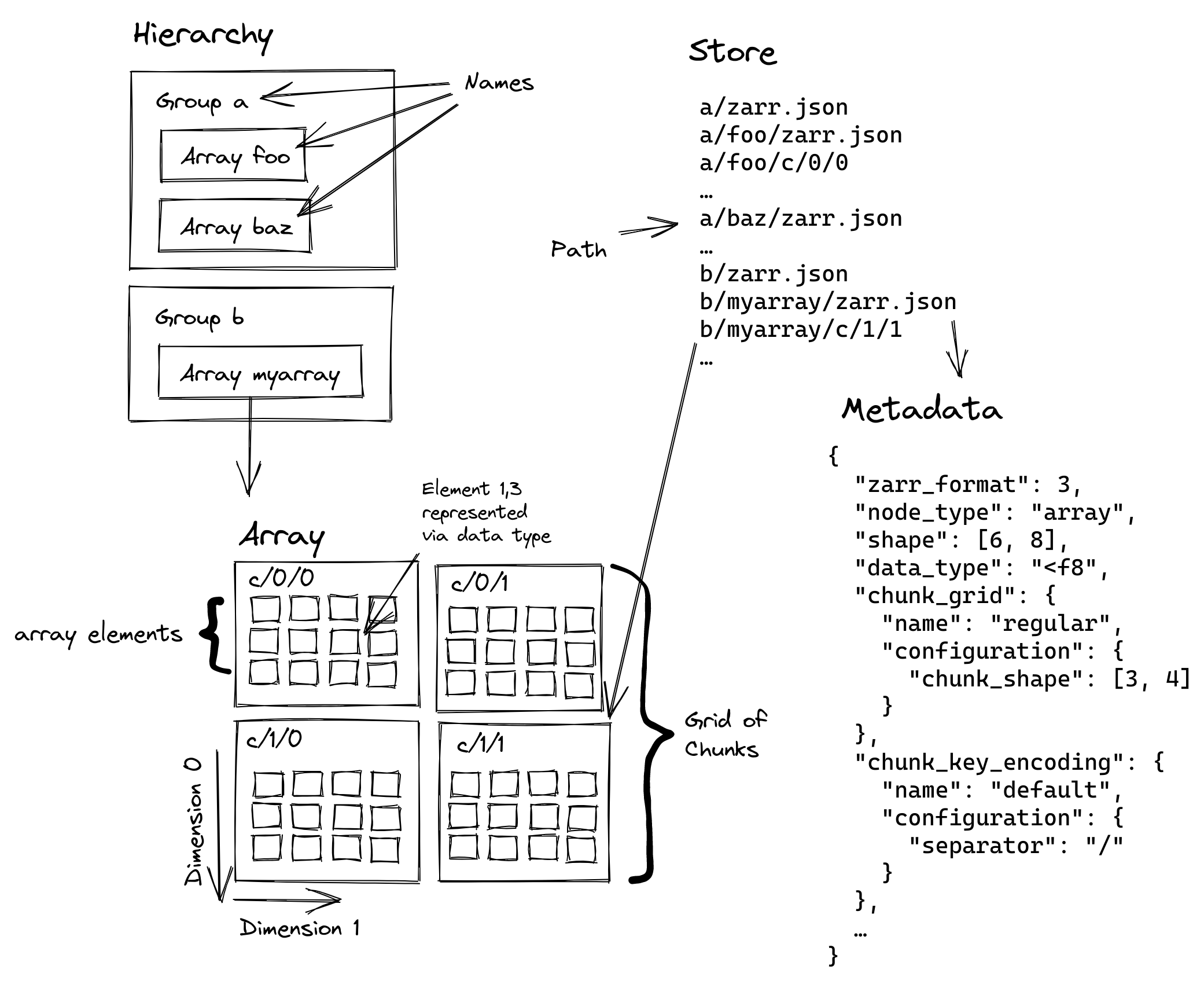
How does zarr relate to xarray?#
Arrays <->
Variables- zarr arrays map well toxarray.VariablesEspecially as zarr v3 includes (optional)
dimension_names
Groups <->
Datasets- zarr groups map reasonably well toxarray.DatasetobjectsOpen a single zarr group in xarray via
xr.open_dataset(store, group='/path', engine='zarr')
Groups must be alignable - But
xarray.Datasets require that all arrays in the Dataset have aligned dimensionsso it is possible to create a zarr group that is not a valid
xarray.Dataset, if the group contains arrays with non-aligning dimensions
No “coordinates” – No arrays are special, so Zarr has no intrinsic concept of “coordinate” vs “data” variables.
So xarray has to save this piece of information as an additional piece of zarr metadata.
Tree of groups <->
DataTree- zarr store has a tree of groupsmaps to either a set of independent
xarray.Datasetsxr.open_groups(store)
or to a single
xarray.DataTreexr.open_datatree(store)xarray.DataTreeenforces alignment between coordinates in parents and child groupsmeans that you could write two
xarray.Datasetsas separate zarr groups that cannot be opened as onexarray.DataTree
coordinate inheritance also means that inherited coordinates are implicitly present on child groups in the
DataTree, but not saved explicitly into each zarr group.
HDF5#
HDF5 (Hierarchical Data Format, version 5) is a general-purpose container for large, heterogeneous, hierarchical data. It includes these core components:
Groups
Nodes in a directed graph that starts at the root /.
They behave like folders in a UNIX filesystem (absolute paths, /sub/group/dataset), and may form cycles or self-links—although most scientific tools avoid that complexity.
Datasets
Rectangular N-dimensional arrays stored inside groups.
Each dimension can optionally carry a dimension scale, an auxiliary dataset that describes the coordinate values along that axis.
Attributes
Small pieces of metadata (strings, scalars, short arrays) attached to the file, any group, or any dataset.
Storage features
Chunking, compression, checksums, parallel I/O via MPI-IO, and more.
These are orthogonal to the logical data model.
NetCDF4#
NetCDF4 builds upon HDF5 - it is really an opinionated subset of HDF5. From the netCDF documentation:
(Note that modifying these files with HDF5 will almost certainly make them unreadable to netCDF-4.)
How does HDF5/netCDF4 relate to xarray?#
Generally very similar - not surprising as netCDF4 was the inspiration for xarray
Similarities#
HDF groups can often be represented by groups in a
DataTreeNetcdf4
Datasetscorrespond to xarrayDatasets(or to groups in theDataTree)NetCDF4 requires that every HDF5 dataset must have a dimension scale attached to each dimension.
These end up working quite similarly to xarray’s dimension coordinates
Differences#
The group structure is not technically a tree - cycles and self-references are allowed in HDF5, unlike in
xarray.DataTree. HDF5 supports links between groups, but xarray does not(Note that this is consistent with UNIX filesystems, which support symbolic links between directories)
NetCDF has an explicit concept of a dimension as a first-class object, which neither HDF5 nor xarray have
means that e.g. a scalar variable can have a dimension in netCDF, but not in xarray
TIFF#
TIFF (Tag Image File Format) is a raster container widely used in biosciences, remote sensing and GIS.
A GeoTIFF is simply a TIFF that stores additional additional georeferencing information tags (CRS, affine transform, etc.) so geospatial software knows where each pixel sits on Earth.
Images (“IFDs”) – each “page” in a TIFF holds a 2-D array of pixels.
Multi-band rasters (e.g. RGB, multi-spectral) appear as separate IFDs or as extra samples within one IFD.
Tags – key–value metadata pairs (datatype, compression, nodata value, CRS, resolution, etc.).
GeoTIFF adds standardised tags like ModelPixelScaleTag, ModelTiepointTag, GeoKeyDirectoryTag.
Compression / tiling – DEFLATE, LZW, etc. Tiling lets software fetch small windows efficiently.
Cloud-optimized GeoTIFF (COG) – same format, arranged so HTTP range requests can stream windows efficiently; xarray handles it transparently when rasterio is compiled with libcurl.
How does TIFF relate to xarray?#
Dimensionality – Each raster image maps well to a single
xarray.Variable, but TIFF is inherently 2-D per band; no native time or vertical axis. If you need 4-D data, NetCDF or Zarr is usually a better fit.No named dimensions - TIFFs don’t have named dimensions for the two axes of the raster.
IFDs as groups - IFDs can be mapped to groups, which may be useful for multi-resolution TIFFs (also known as “overviews”) and multi-page TIFFs.
Metadata depth – single-level tags only (no nested groups). For rich hierarchies, stick to HDF5 / NetCDF-4.
Read – use
rioxarray.open_rasterio()(wraps rasterio) to get an immediate, Dask-chunked DataArray. Howeverrioxarrayis for interacting with GeoTIFFs, not general TIFFs.Write –
DataArray.rio.to_raster("out.tif"); choose compression + tiling via driver_kwargs.
Mapping storage formats to one another#
Note that while we have discussed mapping various file formats to the xarray data model, it is also possible to map different file formats to one another.
For example, the VirtualiZarr library maps a range of file formats (including HDF5 and TIFF) to the Zarr data model, to allow reading data from those formats via the zarr-python API.

